Given a sample table and a design formula, generate a collection of
static plots for exploring the resulting design matrix graphically.
This function is called internally by ExploreModelMatrix(), but
can also be used directly if interactivity is not required.
VisualizeDesign(
sampleData,
designFormula = NULL,
flipCoordFitted = FALSE,
flipCoordCoocc = FALSE,
textSizeFitted = 5,
textSizeCoocc = 5,
textSizeLabsFitted = 12,
textSizeLabsCoocc = 12,
lineWidthFitted = 25,
addColorFitted = TRUE,
colorPaletteFitted = scales::hue_pal(),
dropCols = NULL,
designMatrix = NULL
)Arguments
- sampleData
A
data.frameofDataFramewith sample information.- designFormula
A
formula. All components of the terms must be present as columns insampleData.- flipCoordFitted, flipCoordCoocc
A
logical, whether to flip the coordinate axes in the fitted values/co-occurrence plot, respectively.- textSizeFitted, textSizeCoocc
A
numericscalar giving the text size in the fitted values/co-occurrence plot, respectively.- textSizeLabsFitted, textSizeLabsCoocc
A
numericscalar giving the text size for the axis labels in the fitted values/co-occurrence plot, respectively.- lineWidthFitted
A
numericscalar giving the maximal length of a row in the fitted values plot, before it is split and printed on multiple lines- addColorFitted
A
logicalscalar indicating whether the terms in the fitted values plot should be shown in different colors.- colorPaletteFitted
A
functionreturning a color palette to use for coloring the model coefficients in the fitted values plot.- dropCols
A character vector with columns to drop from the design matrix, or NULL if no columns should be dropped.
- designMatrix
A
numericmatrix, which can be supplied as an alternative todesignFormula. Rows must be in the same order as the rows insampleData.
Value
A list with the following elements:
sampledata: Adata.frame, expanded from the inputsampleDataplotlist: A list of plots, displaying the fitted values for each combination of predictor values, in terms of the model coefficients.designmatrix: The design matrix, after removing any columns indropColspseudoinverse: The pseudoinverse of the design matrixvifs: Adata.framewith calculated variance inflation factorscolors: A vector with colors to use for different model coefficientscooccurrenceplots: A list of plots, displaying the co-occurrence pattern for the predictors (i.e., the number of observations for each combination of predictor values)totnbrrows: The total number of "rows" in the list of plots of fiitted values. Useful for deciding the required size of the plot canvas.
Details
Note that if a design matrix is supplied (via the designMatrix
argument), caution is required in order to interpret especially the
cooccurrence plot in the situation where the provided sampleData
contains additional columns not used to generate the design matrix (or
when it does not contain all the relevant columns).
Examples
VisualizeDesign(
sampleData = data.frame(genotype = rep(c("A", "B"), each = 4),
treatment = rep(c("treated", "untreated"), 4)),
designFormula = ~genotype + treatment
)
#> $sampledata
#> genotype treatment value nSamples
#> 1 A treated (Intercept) 2
#> 2 A untreated (Intercept) + treatmentuntreated 2
#> 3 B treated (Intercept) + genotypeB 2
#> 4 B untreated (Intercept) + genotypeB + treatmentuntreated 2
#>
#> $plotlist
#> $plotlist[[1]]
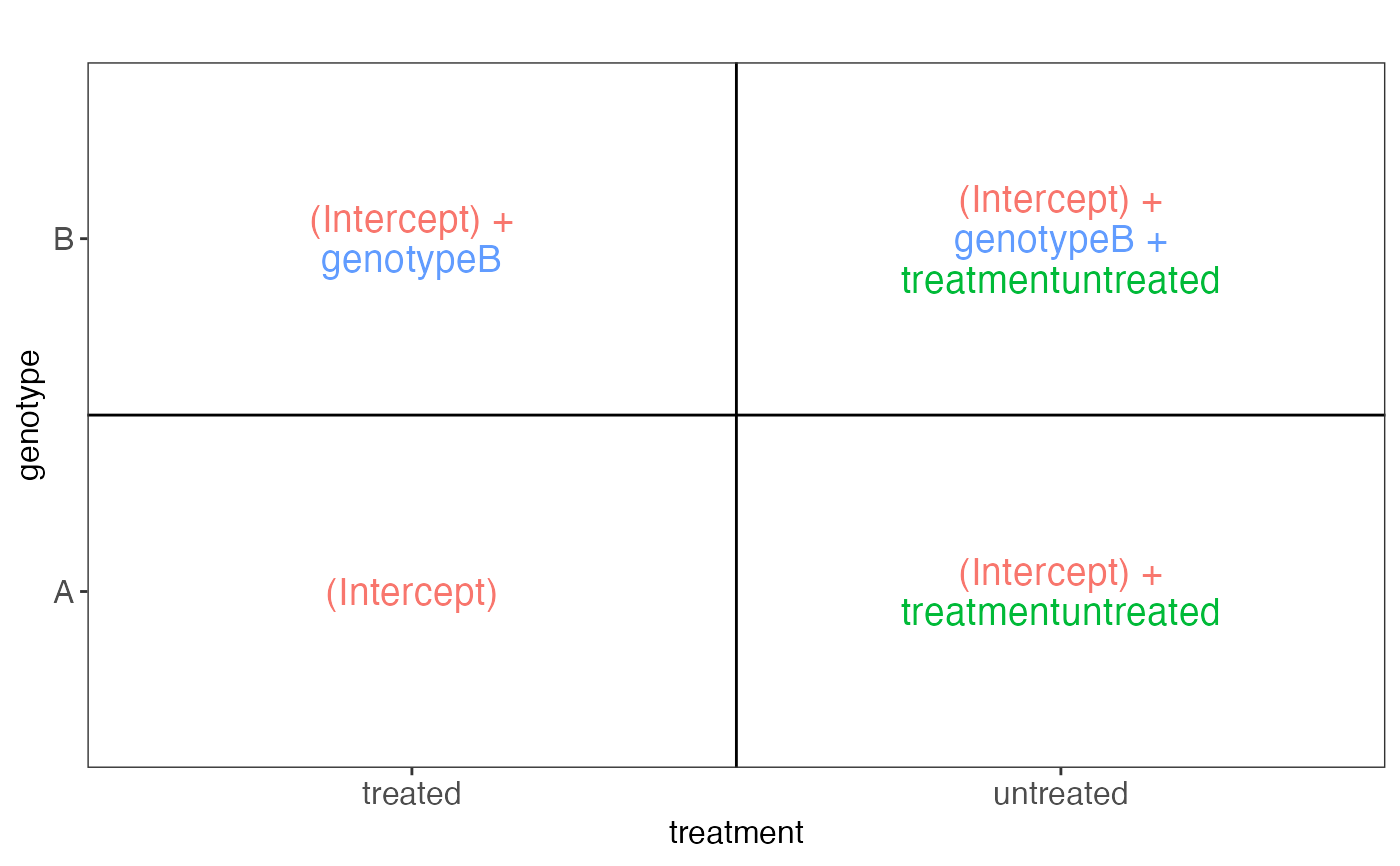 #>
#>
#> $designmatrix
#> (Intercept) genotypeB treatmentuntreated
#> 1 1 0 0
#> 2 1 0 1
#> 3 1 0 0
#> 4 1 0 1
#> 5 1 1 0
#> 6 1 1 1
#> 7 1 1 0
#> 8 1 1 1
#>
#> $pseudoinverse
#> 1 2 3 4 5 6 7 8
#> (Intercept) 0.375 0.125 0.375 0.125 0.125 -0.125 0.125 -0.125
#> genotypeB -0.250 -0.250 -0.250 -0.250 0.250 0.250 0.250 0.250
#> treatmentuntreated -0.250 0.250 -0.250 0.250 -0.250 0.250 -0.250 0.250
#>
#> $vifs
#> coefficient vif
#> 1 genotypeB 1
#> 2 treatmentuntreated 1
#>
#> $colors
#> (Intercept) treatmentuntreated genotypeB
#> "#F8766D" "#00BA38" "#619CFF"
#>
#> $cooccurrenceplots
#> $cooccurrenceplots[[1]]
#>
#>
#> $designmatrix
#> (Intercept) genotypeB treatmentuntreated
#> 1 1 0 0
#> 2 1 0 1
#> 3 1 0 0
#> 4 1 0 1
#> 5 1 1 0
#> 6 1 1 1
#> 7 1 1 0
#> 8 1 1 1
#>
#> $pseudoinverse
#> 1 2 3 4 5 6 7 8
#> (Intercept) 0.375 0.125 0.375 0.125 0.125 -0.125 0.125 -0.125
#> genotypeB -0.250 -0.250 -0.250 -0.250 0.250 0.250 0.250 0.250
#> treatmentuntreated -0.250 0.250 -0.250 0.250 -0.250 0.250 -0.250 0.250
#>
#> $vifs
#> coefficient vif
#> 1 genotypeB 1
#> 2 treatmentuntreated 1
#>
#> $colors
#> (Intercept) treatmentuntreated genotypeB
#> "#F8766D" "#00BA38" "#619CFF"
#>
#> $cooccurrenceplots
#> $cooccurrenceplots[[1]]
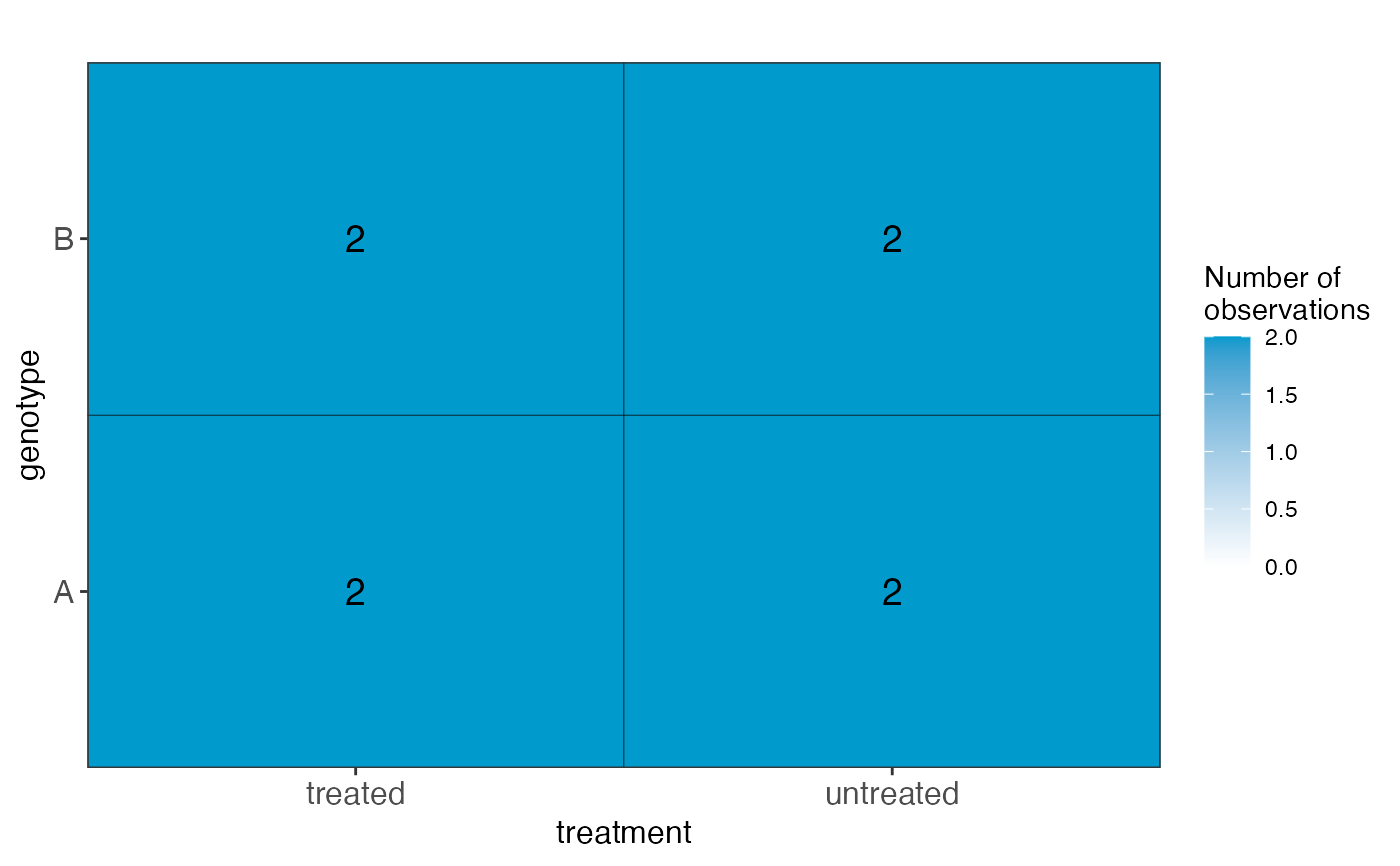 #>
#>
#> $totnbrrows
#> [1] 2
#>
#>
#>
#> $totnbrrows
#> [1] 2
#>